| Issue |
Ann. Limnol. - Int. J. Lim.
Volume 57, 2021
|
|
|---|---|---|
| Article Number | 7 | |
| Number of page(s) | 7 | |
| DOI | https://doi.org/10.1051/limn/2021005 | |
| Published online | 08 March 2021 | |
Research Article
Genetic diversity and population structure of Hemibagrus guttatus (Bagridae, Siluriformes) in the larger subtropical Pearl River based on COI and Cyt b genes analysis
1
Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou 510380, PR China
2
Université de Toulouse - Paul Sabatier, 118 route de Narbonne, 31062, Toulouse Cedex, France
* Corresponding author: shuai6662000@aliyun.com
Received:
28
July
2020
Accepted:
23
January
2021
Understanding the genetic diversity and population structure of fish species is crucial for the sustainable use and protection of fish germplasm resources. Hemibagrus guttatus (Bagridae, Siluriformes) is widely distributed in the large subtropical Pearl River (China) and is commercially important. It's population have been declining. The genetic diversity of wild H. guttatus is not clear, despite its important ecological significance. In this paper, genes mitochondrial cytochrome c oxidase subunit I (COI) and cytochrome b (Cyt b) were used to analyze the genetic structure of H. guttatus population collected from six geographical populations in the main streams of the Pearl River. The results showed that the nucleotide diversity (π) and haplotype diversity (Hd) of wild H. guttatus was low (π < 0.005; Hd < 0.5). In addition, H. guttatus haplotypes did not cluster into clades according to geographical distribution, as revealed by neighbor-joining tree analysis. Analysis of molecular variance analysis (AMOVA) and F-statistics (Fst) values showed high homogeneity among wild H. guttatus populations. Our results suggest that there is degradation in germplasm resources of H. guttatus that could destabilize the sustainable use of this species and there was an urgent need for conservation of this species in South China.
Key words: Pearl River / Hemibagrus guttatus / genetic diversity / COI gene / Cyt b gene
© EDP Sciences, 2021
1 Introduction
The Pearl River is a large subtropical river in southern China, stretching some 2400 km. It originates from Maxiong Mountain of Yunnan province, and flows into the South China Sea. This river is not only a hot spot of global biodiversity research, but is also an important gene pool of aquatic biological resources, due to its high habitat heterogeneity (Shuai et al., 2017). However, due to human disturbances, such as over-fishing and the construction of dams in recent years, fish migratory paths have been obstructed and spawning grounds have disappeared, which has resulted in a sharp degradation of fish germplasm resources. Hemibagrus guttatus belongs to the family Bagridae fishes, they are benthic and settled fishes with relatively weak migration capability (Chu et al., 1999). This kind of lifestyle make the long distance migration hard to realize for them, therefore, H. guttatus is not a wildly distribute species and mainly found in the Pearl River basin. H. guttatus is a commercially important fish in southern China and has earned a reputation as the “king of freshwater fish” due to its high nutritional value (Kottelat, 2001). Nevertheless, in recent decades, H. guttatus population declined sharply due to over-fishing, water pollution, and hydraulic constructions and led to a degradation of their germplasm resources. Therefore, it is particularly important to study the genetic diversity of wild H. guttatus in the Pearl River (World Commission on Dams, 2000).
However, up to now, only a few studies on the genus Hemibagrus have been conducted, such as the study of diversity and evolutionary rate of bagrid catfish (including Hemibagrus, Pseudobagrus, Pelteobagrus and Leiocassis) (Peng et al., 2002; Ku et al., 2007), the research of the phylogeny and biogeography of H. guttatus (Yang and He, 2008) and the work of genetic diversity of Hemibagrus macropterus populations (Yang et al., 2009). The current genetic diversity of H. guttatus in the river basins of southern China is still unknown, despite its important ecological significance and its significance for conservation of fishery resources.
Mitochondrial genes are maternally inherited (Haye et al., 2010). The evolutionary rates of mitochondrial cytochrome c oxidase subunit I (COI) and mitochondrial cytochrome b (Cyt b) genes are relatively moderate (Subramanian et al., 2009), which makes them ideal molecular markers for the study of intraspecific genetic diversity in aquatic organisms (Beenaerts et al., 2010; Haye et al., 2010; Sun et al., 2012). The monitoring of germplasm resources of the wildlife in the Pearl River basin should focus on the genetic level, instead of the number of individuals. Previous study have reported that the H. guttatus haplotypes identified base on Cyt b gene from the Wast river, North river and East River were very common and similar, indicated that H. guttatus from the Pearl River System was highly homogeneous (Yang et al., 2008). In this study, we combined the two genes (COI gene and Cyt b gene), with longer Nucleotide base sequence, to improve the sensitivity to analysis the genetic diversity of H. guttatus.
The purpose of this study was to understand the genetic diversity and population structure of the H. guttatus in the Pearl River. Our results will provide a scientific basis for the conservation and management of commercial fish germplasm resources in the Pearl River.
2 Materials and methods
2.1 Sampling
In this study, a total of six sampling sites were set up, covering almost the entire mainstream of the Pearl River. The average interval between sampling sites was about 200 km. Detailed information and distribution of the sampling sites are shown in Table 1 and Figure 1. A total of 127 H. guttatus individuals were collected by hook fishing. The pectoral fins or tail fins were cut for COI and Cyt b genes analysis.
Sampling information of H. guttatus.
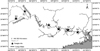 |
Fig. 1 Sampling locations of H. guttatus. |
2.2 DNA extraction, amplification, and sequencing
Fin tissues (2 mg) were cut into fine pieces for total DNA extraction, using standard phenol/chloroform extraction method (Sambrook et al., 2001). A 606 bp fragment at the 5' end of the mitochondrial COI gene was amplified using the universal primers:
FishF: 5'-TCAACCAACCACAAAGACATTGGCAC-3'; and
FishR: 5'-TAGACTTCTGGGTGGCCAAAGAATCA-3'.
A 1005 bp fragment at the 5' end of the mitochondrial Cyt b gene was amplified using the universal primers:
L14724: 5'-GACTTGAAAAACCACCGTTG-3'; and
H15915: 5'-CTCCGATCTCCGGATTACAAGAC-3' (Xiao et al., 2001; Sun et al., 2012).
The PCR reaction was prepared for a final volume of 40 uL, containing 4 uL of 10X buffer, 2 uL of dNTPs (10 mM), 1 uL each of upstream and downstream primers (20 uM), 2 uL of Taq polymerase (5 U), 2 uL of genomic DNA as template; ultrapure water was added to the total volume of 40 uL. The PCR program was as follows: pre-denaturation at 95 °C for 3 min, then 35 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C for 30 s, and extension at 72 °C for 1 min, and then a final extension at 72 °C for 10 min. Amplification products were analyzed by 1% agarose gel electrophoresis and sent to Guangzhou IGE Biotechnology Ltd for sequencing. Sequencing primers were same as the amplification primers.
2.3 Data analysis
Seqman software was used to align the forward and reverse sequences. All the sequences were multiply aligned by Clustal X software (Thompson et al., 1997). The sequences nucleotide composition was analyzed using MEGA 7.0 software (Tamura et al., 2011). The genetic distance was calculated based on the Kimura two-parameter (K2P) model. Phylogenetic trees of individuals and haplotypes were constructed using the neighbor-joining (NJ) method (Nei and Kumar, 2000), whose confidence was tested by 1000 bootstrap replicates. In addition, a median-joining network of all haplotypes was constructed using Network 4.6 software (www. fluxus-engineering.com). Arlequin 3.5 software was used to estimate the genetic differentiation coefficient (FST) between populations based on a pairwise difference model (Garcia et al., 2003; Excoffier and Lischer, 2010). Analysis of molecular variance (AMOVA) was performed on populations of H. guttatus at two different scales using Arlequin software. For one scale, the six populations of H. guttatus were combined into one group to verify if there were significant genetic differences among the six populations. For the other scale, the six populations were divided into three groups according to their geographical location (upstream, middle stream, and downstream), to verify whether there were significant genetic differences among the three groups.
3 Results
3.1 Information of base composition and variable sites
A total of 127 H. guttatus individuals from six geographical populations in the mainstream of the Pearl River were analyzed based on a 1611 bp sequences, which included a 606 bp fragment at the 5' end of the mitochondrial COI gene, and a 1005 bp fragment at the 5' end of mitochondrial Cyt b gene. The average content of A, T, G and C in the COI and the Cyt b gene sequence was 28.6%, 31.5%, 14.9% and 25.1%, respectively; which shows an AT bias (AT 60.1%, GC 39.9%). The haplotype number in all populations was 15. The haplotype diversity index was 0.465 ± 0.054. The nucleotide diversity index was 0.00042 ± 0.00007. The indexes of genetic diversity of each geographical population are shown in Table 2.
Genetic diversity indexes in H. guttatus populations based on COI and Cyt b gene sequences.
3.2 Haplotype distribution and haplotype analysis of COI and Cyt b genes sequence
No insertions or deletions were found in the COI and Cyt b gene sequences. There were 19 mutation sites, including 12 single mutation sites and seven parsimony-informative mutation sites. Mutation sites 544, 1591, 1366, and 1399 were at the first position of the codons, mutation sites 320 and 692 were at the second position of the codon, and the rest of the mutation sites were at the third position of the codons (Tab. 3).
Genetic structure analysis showed that the bootstrap values for most node branches in the NJ phylogenetic tree of haplotypes were higher than 50%. The haplotypes of populations were widely distributed in the NJ phylogenetic tree of haplotypes without a structure that reflected geographical distribution (Fig. 2).
Nineteen polymorphic sites, defined 15 haplotypes, were found on the COI and Cyt b gene sequences. Haplotype H1 occurred most frequently, accounting for 92.97% of all individuals; it was found in the six geographical populations. The second most common haplotype was H3, it was found in all the geographical populations, except in Heshan. Haplotypes, H2, H5–7, H9–11, and H13–15 were unique to single geographical populations. The distribution of each haplotype by geographical location and the network of the 15 haplotypes are shown in Figure 3 and Table 4.
Haplotypes of COI and Cyt b gene sequences in H. guttatus populations.
 |
Fig. 2 Neighbor-joining tree of haplotypes based on COI and Cyt b gene sequences. |
 |
Fig. 3 Median-joining haplotypes network of H.guttatus based on COI and Cyt b gene sequences. Circle areas are proportional to haplotype frequencies, while colored portions represent the proportions of the same haplotype that occurs in each sampling region. |
Distribution of haplotypes in H. guttatus populations.
3.3 Population differentiation
The pairwise FST values between populations ranged from −0.01410 to 0.20212, based on the COI and Cyt b gene sequences. The pairwise differences between the population of Zhaoqing and the other five populations were significant (p < 0.05). The pairwise differences between the rest of the geographical populations were not significant (p > 0.05), indicating that high genetic homogeneity existed among the opulations (Tab. 5). AMOVA analysis showed no obvious differentiation among the three groups (p = 0.20821) (Tab. 6).
Genetic differentiation coefficient (FST Values) between wild H. guttatus populations based on COI and Cyt b sequences.
AMOVA analysis of H. guttatus populations based on COI and Cyt b 15 sequences.
3.4 Populations' history
Tajima'D and Fu's Fs teat results are shown in Table 7. For Tajima'D, Baiceng, Heshan and Tengxian were significant D values (p < 0.05). For Fu's Fs, all of the populations were significant negative Fs values (p < 0.05) except Heshan and Zhaoqing. Overall, the neutral teat results showed not significant negative values. The results of the mismatch distribution analysis are shown in Figure 4. Combined with the results of the neutrality test, it is speculated that the population may not have had any population expansion in history.
Neutrality test of H. guttatus populations based on COI and Cyt b sequences.
 |
Fig. 4 Mismatch distribution of pairwise difference of COI and Cyt b gene sequences in H. guttatus. |
4 Discussions
In this study, the genetic diversity and population structure of H. guttatus in the Pearl River was investigated based on mitochondrial COI gene and Cyt b gene sequences. 15 haplotypes and 19 polymorphic sites were detected in a 1611 bp long segment of COI gene and Cyt b gene sequences from 127 individuals collected from six locations. The Hd and nucleotide diversity (π) were 0.465 and 0.00042 respectively. The haplotype diversity and the nucleotide diversity in our study were lower than in other studies on aquatic organisms, such as Portunus sanguinolentus (h = 0.9576, π = 0.0051) along south China Sea and east China Sea (Ren et al., 2016); the bighead carp (Aristichthys nobilis) (h = 0.9083, π = 0.0032) in China (Li et al., 2010); and Penaeus monodon (h = 0.927, π = 0.0294) in Thailand (Khamnamtong et al., 2009). Such a low level of genetic diversity may be the result of human activities, such as the construction of dams. At present, 32 hydropower stations with capacities larger than 100 MW have been built in the Pearl River. These hydropower stations not only affected spawning grounds, but also reduced the exchange of genes between different populations, resulting in a sharp decrease in the population number and genetic diversity (Wang et al., 2004; Tan et al., 2010; Shuai et al., 2017). In addition to hydropower stations, overfishing has led to a genetic bottleneck that also cannot be ignored (Khamnamtong et al., 2009).
The evolutionary events, such as genetic drift, migration and natural selection, may also play a role in determining the patterns of genetic variation (Rubinoff, 2006; Yamaguchi et al., 2010). The not significant FST values indicate a higher homogenization among populations. Male H. guttatus need six years to reach the sexual maturity while female fish need seven years. Longer H. guttatus generations lead a lower evolutionary rate (Wang et al., 2017), which makes H. guttatus population differentiation slower when compared with carps.
Previous studies indicated that glacial cycles have a significant impact on the genetic diversity and distribution of many existing species. However, different species have different responses to glacial cycles (Hewitt, 2000, 2004; Provan and Bennett, 2008). In our study, neutral tests and mismatch analysis showed that H. guttatus populations in the Pearl River basin have maintained a relatively stable population size in the past, and have not experienced population expansion. This is mainly due to the fact that the Pearl River basin is located in the tropical and subtropical regions of southern China, which have mild climate (Weaver et al., 1998; Ju et al., 2007). Fish species can maintain a stable niche during a glacial period, which maintains a relatively stable population. In the recent years, the population size of H. guttatus have been declined massively because of the overfishing, which maybe was the main reason that lead the genetic diversity of the H. guttatus populations shown the relatively lower diversity in the Pearl River basin.
In conclusion, low levels of genetic variation in the populations of H. guttatus in the Pearl River and low sequence diversity among the six populations were revealed. Understanding current situation such as those outlined in this research is the basis of conservation of H. guttatus population diversity, which is a critical resource for successful sustainable fishery in the Pearl River.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (General Program NO. 31870527) and the China-ASEAN Maritime Cooperation Fund (No: CAMC-2018F).
References
- Beenaerts N, Pethiyagoda R, Ng PKL, Yeo DC, Bex GJ, Bahir MM, Artois T. 2010. Phylogenetic diversity of Sri Lankan freshwater crabs and its implications for conservation. Mol Ecol 19: 183–196. [PubMed] [Google Scholar]
- Chu XL, Zheng BS, Dai DY. 1999. Fauna Sinica, Class Teleostei, Siluriformes (in Chinese). Beijing: Scientific Press. [Google Scholar]
- Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to Perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567. [Google Scholar]
- Garcia BA, Manfredi C, Fichera L, Segura EL. 2003. Short report: variation in mitochondrial 12s and 16s ribosomal DNA sequences in natural populations of Triatoma infestans (Hemiptera; Reduviidae). Am J Trop Med Hyg 68: 692–694. [PubMed] [Google Scholar]
- Haye PA, Salinas P, Acuña E, Poulin E. 2010. Heterochronic phenotypic plasticity with lack of genetic differentiation in the southeastern Pacific squat lobster Pleuroncodes monodon . Evol Dev 12: 628–633. [PubMed] [Google Scholar]
- Hewitt GM. 2000. The genetic legacy of the quaternary ice ages. Nature 405: 907–913. [CrossRef] [PubMed] [Google Scholar]
- Hewitt GM. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philos T R Soc B 359: 183–195. [Google Scholar]
- Ju L, Wang H, Jiang D. 2007. Simulation of the last glacial maximum climate over East Asia with a regional climate model nested in a general circulation model. Palaeogeogr Palaeoclimatol Palaeoecol 248: 376–390. [Google Scholar]
- Khamnamtong B, Klinbunga S, Menasveta P. 2009. Genetic Diversity and Geographic Differentiation of the Giant Tiger Shrimp (Penaeus monodon) in Thailand Analyzed by Mitochondrial COI Sequences. Biochem Genet 47: 42–45. [PubMed] [Google Scholar]
- Kottelat M. 2001. Fishes of Laos. Colombo: WHT Publications. [Google Scholar]
- Ku XY, Peng ZG, Diogo R, He SP. 2007. MtDNA phylogeny provides evidence of generic polyphyleticism for East Asian bagrid catfishes. Hydrobiologia 579: 147–159. [Google Scholar]
- Li SF, Yang QL, Xu JW, Wang CH, Duane CC, Lu GQ. 2010. Genetic Diversity and Variation of Mitochondrial DNA in Native and Introduced Bighead Carp. Trans Am Fish Soc 4: 937–946. [Google Scholar]
- Nei M, Kumar S. 2000. Molecular Evolution and Phylogenetics. New York: Oxford University Press. [Google Scholar]
- Peng ZG, He SP, Zhang YG. 2002. Mitochondrial cytochrome b sequence variations and phylogeny of the East Asian bagrid catfishes. Prog Nat Sci 12: 421–425. [Google Scholar]
- Provan J, Bennett K. 2008. Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol 23: 564–571. [PubMed] [Google Scholar]
- Ren GJ, Ma HY, Ma CY, Wang W, Chen W, Ma LB. 2016. Genetic diversity and population structure of Portunus sanguinolentus (Herbst, 1783) revealed by mtDNA COI sequences. Mitochondrial DNA A 28: 740–746. [Google Scholar]
- Rubinoff D. 2006. Utility of mitochondrial DNA barcodes in species conservation. Cons Biol 20: 1026–1033. [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. 2001. Molecular Cloning: a Laboratory Manual, third ed. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shuai FM, Li XH, Chen FC, Li YF, Lek S. 2017. Spatial patterns of fish assemblages in the Pearl River, China: environmental correlates. Fund Appl Limnol 189: 329–340. [Google Scholar]
- Subramanian S, Denver DR, Millar CD, Heupink T, Aschrafi A, Emslie SD, Baroni C, Lambert DM. 2009. High mitogenomic evolutionary rates and time Dependency. Trends Genet 25: 482–486. [PubMed] [Google Scholar]
- Sun P, Shi Z, Yin F, Peng S. 2012. Population genetic structure and demographic history of Pampus argenteus in the Indo-West Paci c inferred from mitochondrial cytochrome b sequences. Biochem Syst Ecol 43: 54–63. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [CrossRef] [PubMed] [Google Scholar]
- Tan XC, Li XH, Lek S, Li YF, Wang C, Li J, Luo JR. 2010. Annual dynamics of the abundance of fish larvae and its relationship with hydrological variation in the Pearl River. Environ Biol Fish 88: 217–225. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [CrossRef] [PubMed] [Google Scholar]
- Wang ZW, Wu QJ, Zhou JF, Ye YZ. 2004. Geographic Distribution of Pelteobagrus fulvidraco and Pelteobagrus vachelli in the Yangtze River Based on Mitochondrial DNA Markers. Biochem Genet 42: 391–400. [PubMed] [Google Scholar]
- Wang W, Ma CY, Chen W, Zhang H, Kang W, Ni Y, Ma LB. 2017. Population genetic diversity of Chinese sea bass (Lateolabrax maculatus) from southeast coastal regions of China based on mitochondrial COI gene sequences. Biochem Syst Ecol 71: 114–120. [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. 1998. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469. [PubMed] [Google Scholar]
- World Commission on Dams (WCD). 2000. Dams and development: a new framework for decision-making. London: Earthscan. [Google Scholar]
- Xiao WH, Zhang YP, Liu HZ. 2001. Molecular systematic of Xenocyprinae (Teleostei: Cyprinidae): taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol Phylogenet Evol 18: 163–173. [Google Scholar]
- Yamaguchi K, Nakajima M, Taniguchi N. 2010. Loss of genetic variation and increased population differentiation in geographically peripheral populations of Japanese char Salvelinus leucomaenis . Aquaculture 308: S20–S27. [Google Scholar]
- Yang L, He S. 2008. Phylogeography of the freshwater catfish Hemibagrus guttatus (Siluriformes, Bagridae): Implications for South China biogeography and influence of sea-level changes. Mol Phylogenet Evol 49: 393–398. [Google Scholar]
- Yang L, Mayden RL, He S. 2009. Population genetic structure and geographical differentiation of the Chinese catfish Hemibagrus macropterus (Siluriformes, Bagridae): Evidence for altered drainage patterns. Mol Phylogenet Evol 51: 405–411. [Google Scholar]
Cite this article as: Kuang T, Shuai F, Li X, Chen W, Lek S. 2021. Genetic diversity and population structure of Hemibagrus guttatus (Bagridae, Siluriformes) in the larger subtropical Pearl River based on COI and Cyt b genes analysis. Ann. Limnol. - Int. J. Lim. 57: 7
All Tables
Genetic diversity indexes in H. guttatus populations based on COI and Cyt b gene sequences.
Genetic differentiation coefficient (FST Values) between wild H. guttatus populations based on COI and Cyt b sequences.
All Figures
 |
Fig. 1 Sampling locations of H. guttatus. |
| In the text | |
 |
Fig. 2 Neighbor-joining tree of haplotypes based on COI and Cyt b gene sequences. |
| In the text | |
 |
Fig. 3 Median-joining haplotypes network of H.guttatus based on COI and Cyt b gene sequences. Circle areas are proportional to haplotype frequencies, while colored portions represent the proportions of the same haplotype that occurs in each sampling region. |
| In the text | |
 |
Fig. 4 Mismatch distribution of pairwise difference of COI and Cyt b gene sequences in H. guttatus. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


